SeqUnwinder results (simulateOverlap_sequnwinder_05092017)
SeqUnwinder version 0.1.2 run completed on: 2017/05/09 18:44:29 with arguments:
--out simulateOverlap_sequnwinder_05092017 --threads 10 --debug --memepath /Users/akshaykakumanu/Softwares/src/bin/bin/ --geninfo mm10.info --seq /Users/akshaykakumanu/genome/mm10.fa --genseqs simulateOverlap.seqs --win 150 --mink 4 --maxk 5 --r 10 --x 3 --maxscanlen 15
Error on training data
| Class |
TP Rate |
FP Rate |
Precision |
Recall |
F-Measure |
MCC |
ROC Area |
PRC Area |
| X#B
| 0.725 |
0.067 |
0.683 |
0.725 |
0.704 |
0.643 |
0.935 |
0.736 |
| Y#A
| 0.628 |
0.063 |
0.665 |
0.628 |
0.646 |
0.578 |
0.911 |
0.712 |
| X#A
| 0.645 |
0.074 |
0.637 |
0.645 |
0.641 |
0.568 |
0.906 |
0.718 |
| Y#C
| 0.673 |
0.055 |
0.708 |
0.673 |
0.69 |
0.63 |
0.911 |
0.757 |
| X#C
| 0.746 |
0.05 |
0.749 |
0.746 |
0.748 |
0.697 |
0.951 |
0.803 |
| Y#B
| 0.626 |
0.082 |
0.604 |
0.626 |
0.615 |
0.536 |
0.896 |
0.678 |
Stratified cross-validation
| Class |
TP Rate |
FP Rate |
Precision |
Recall |
F-Measure |
MCC |
ROC Area |
PRC Area |
| X#B |
0.378 |
0.083 |
0.475 |
0.378 |
0.421 |
0.324 |
0.801 |
0.442 |
| Y#A |
0.499 |
0.075 |
0.572 |
0.499 |
0.533 |
0.448 |
0.846 |
0.579 |
| X#A |
0.609 |
0.118 |
0.509 |
0.609 |
0.555 |
0.459 |
0.862 |
0.575 |
| Y#C |
0.635 |
0.085 |
0.598 |
0.635 |
0.616 |
0.536 |
0.88 |
0.647 |
| X#C |
0.497 |
0.076 |
0.568 |
0.497 |
0.53 |
0.445 |
0.857 |
0.578 |
| Y#B |
0.583 |
0.123 |
0.487 |
0.583 |
0.531 |
0.429 |
0.857 |
0.551 |
Weighted K-mer models.
De novo motifs
Label specific scores of de novo motifs
| Extended version |
Compact version |
 |
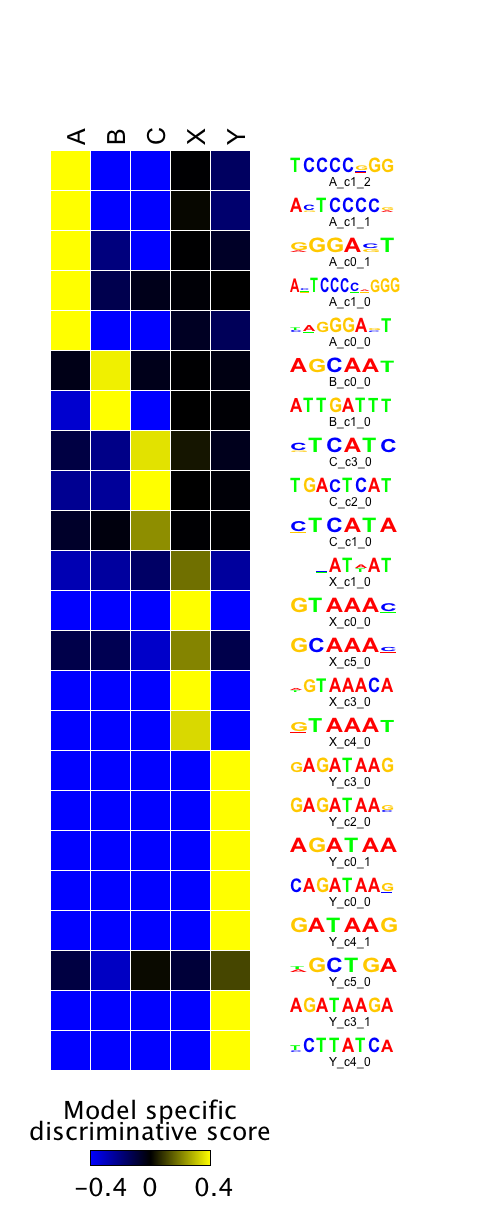 |
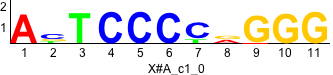
Discriminative motifs. Try inputting these motifs into STAMP for validation.
 rc
rc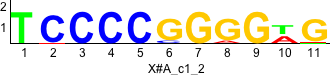 rc
rc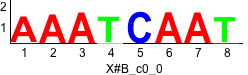 rc
rc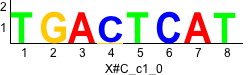 rc
rc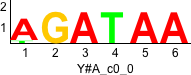 rc
rc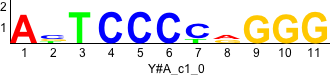 rc
rc rc
rc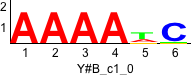 rc
rc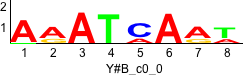 rc
rc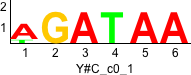 rc
rc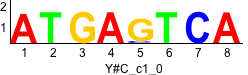 rc
rc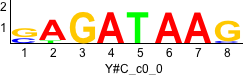 rc
rc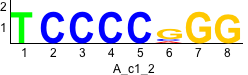 rc
rc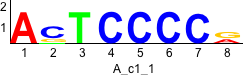 rc
rc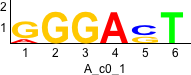 rc
rc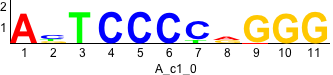 rc
rc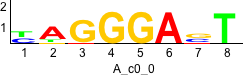 rc
rc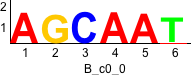 rc
rc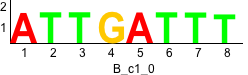 rc
rc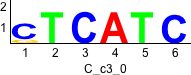 rc
rc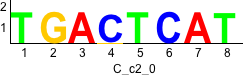 rc
rc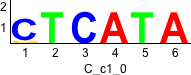 rc
rc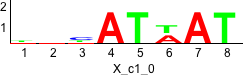 rc
rc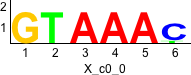 rc
rc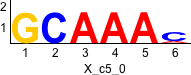 rc
rc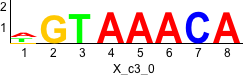 rc
rc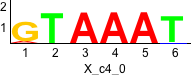 rc
rc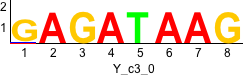 rc
rc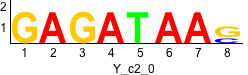 rc
rc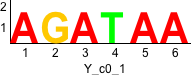 rc
rc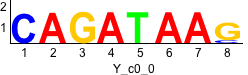 rc
rc rc
rc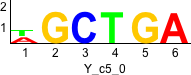 rc
rc rc
rc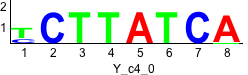 rc
rc rc
rc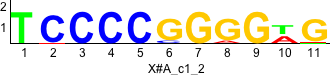 rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc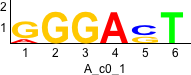 rc
rc rc
rc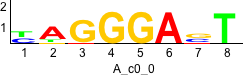 rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc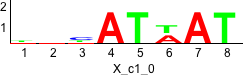 rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc rc
rc
