 rc
rc rc
rc rc
rcMultiGPS version 0.5 run completed on: 2014/03/03 04:05:51 with arguments: --geninfo genomes/mm9/mm9.info --threads 4 --exclude mm9_excludes.txt --design cdx2.design --verbose --probshared 0.99 --memepath software/meme_4.9.0/bin --mememinw 6 --mememaxw 16 --seq genomes/mm9 --out Cdx2
| Condition | Events | File | Positional Prior Motif | Motif Relative Offset |
|---|---|---|---|---|
| ES | 5098 | Cdx2_ES.events |  rc rc |
5 |
| pMN | 39832 | Cdx2_pMN.events |  rc rc |
4 |
| Endo | 37824 | Cdx2_Endo.events |  rc rc |
5 |
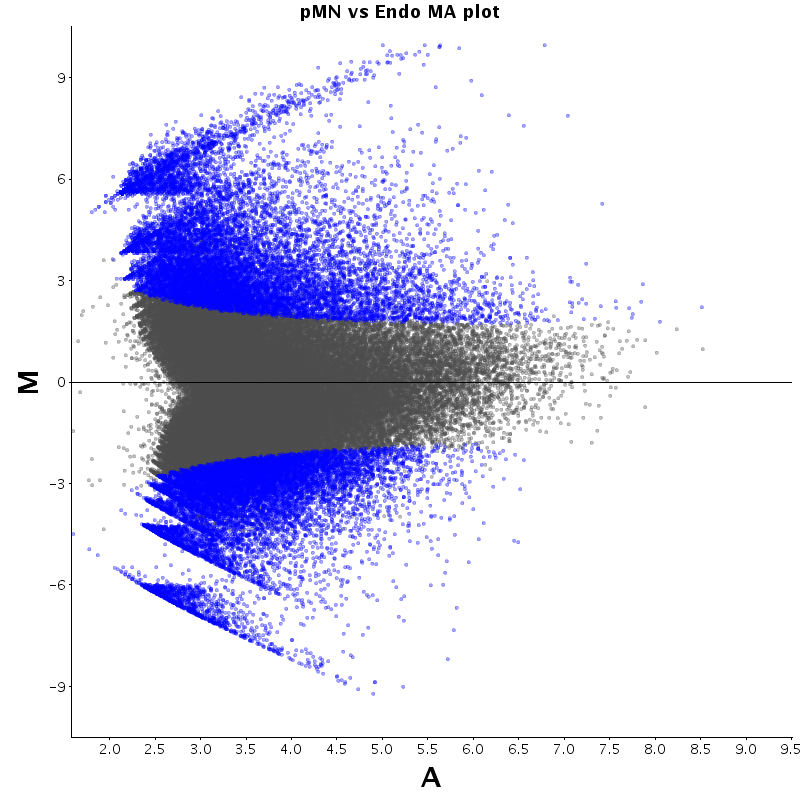
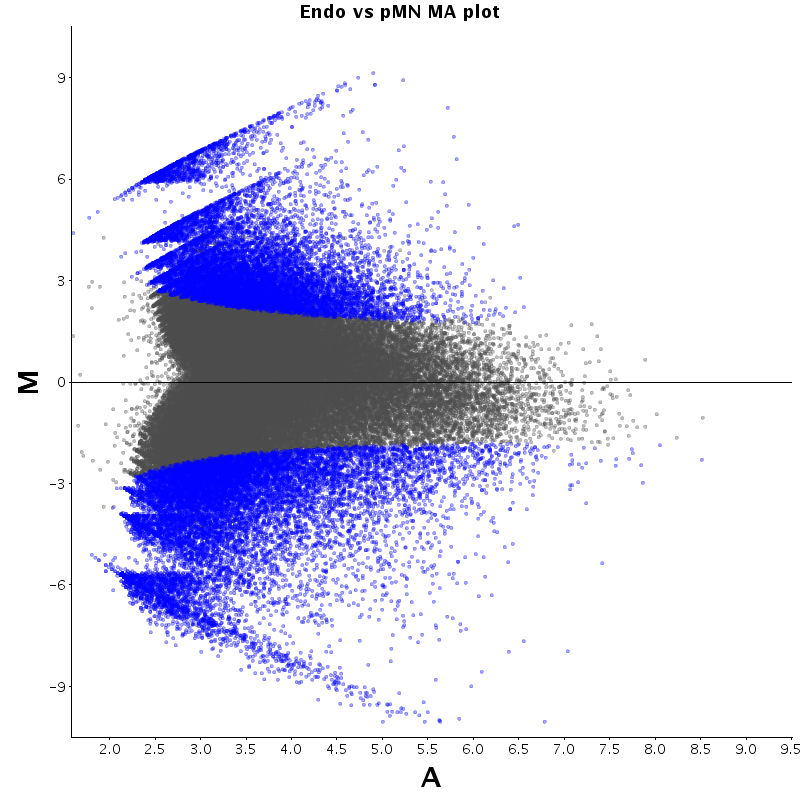
The table displays counts of events that are significantly enriched in the row condition with respect to the column condition. Click on the counts to get the differential event data file.
| Diff | ES | pMN | Endo |
|---|---|---|---|
| ES | - | 2530 | 2727 |
| pMN | 12383 | - | 9320 |
| Endo | 13764 | 8896 | - |
| Diff | ES | pMN | Endo |
|---|---|---|---|
| ES | 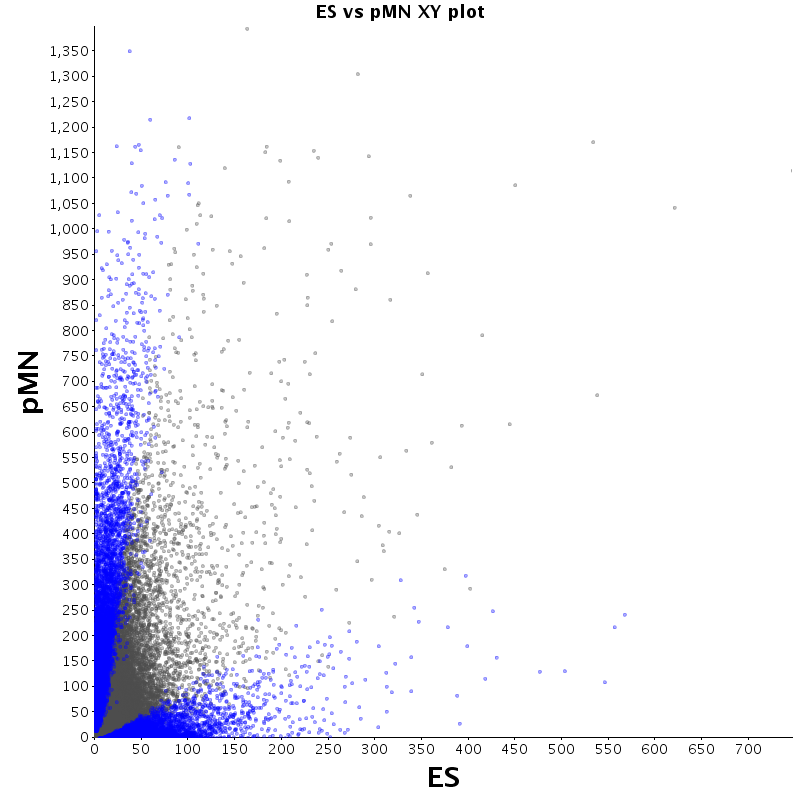 |
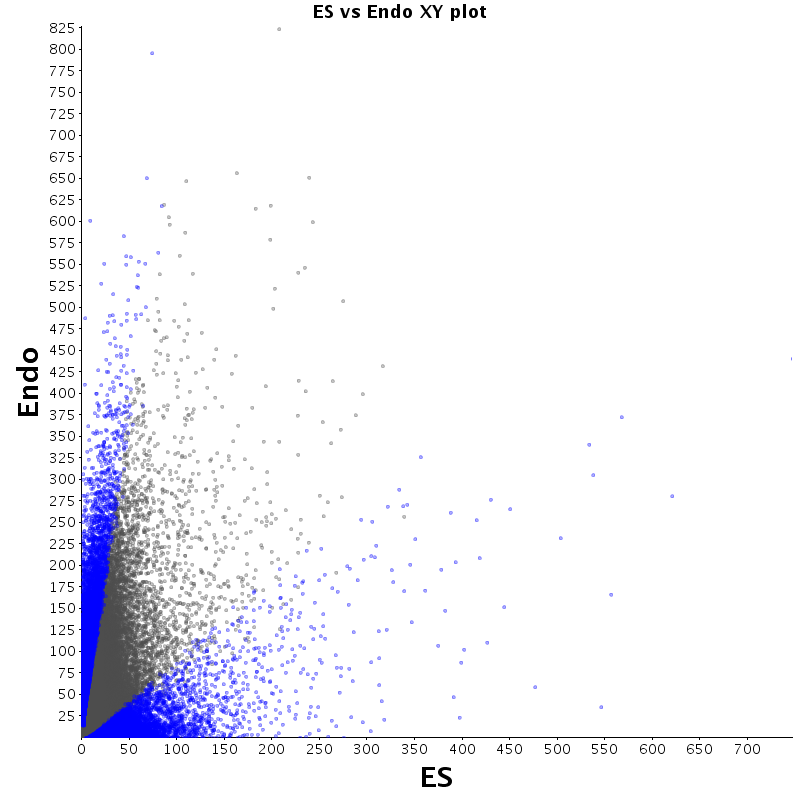 |
|
| pMN | 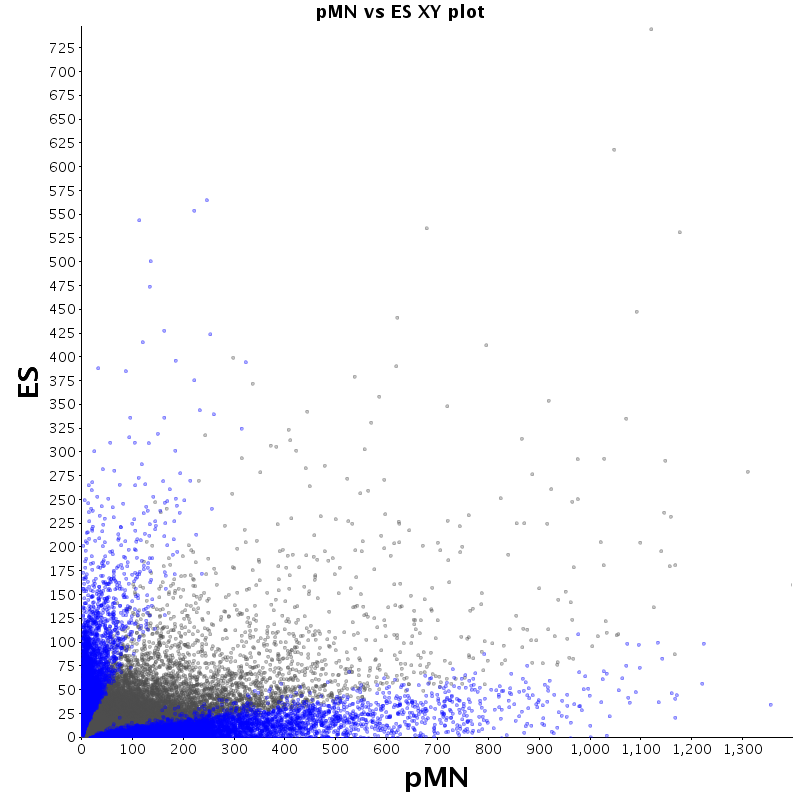 |
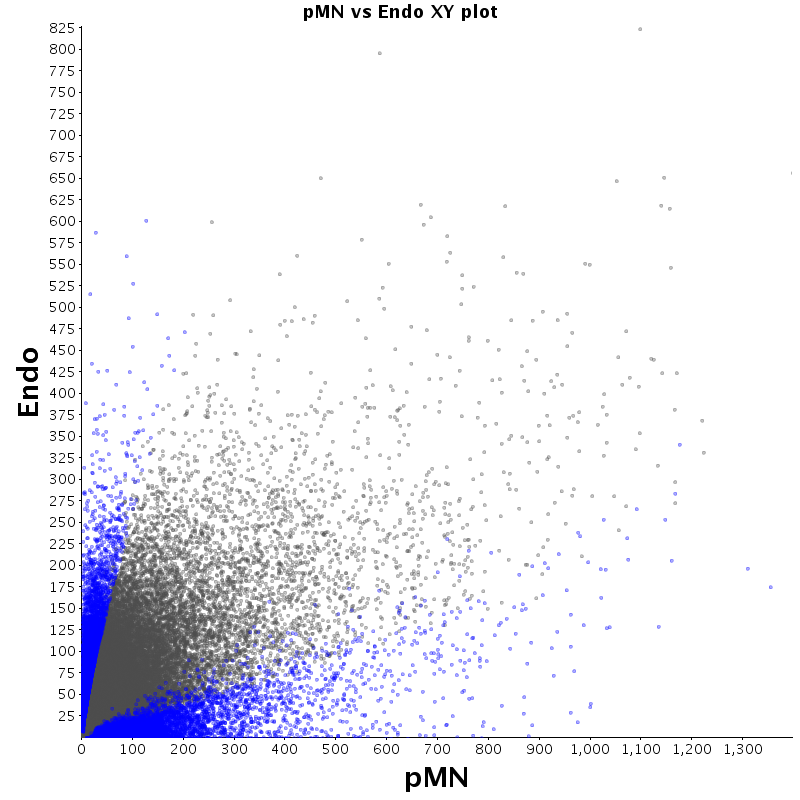 |
|
| Endo | 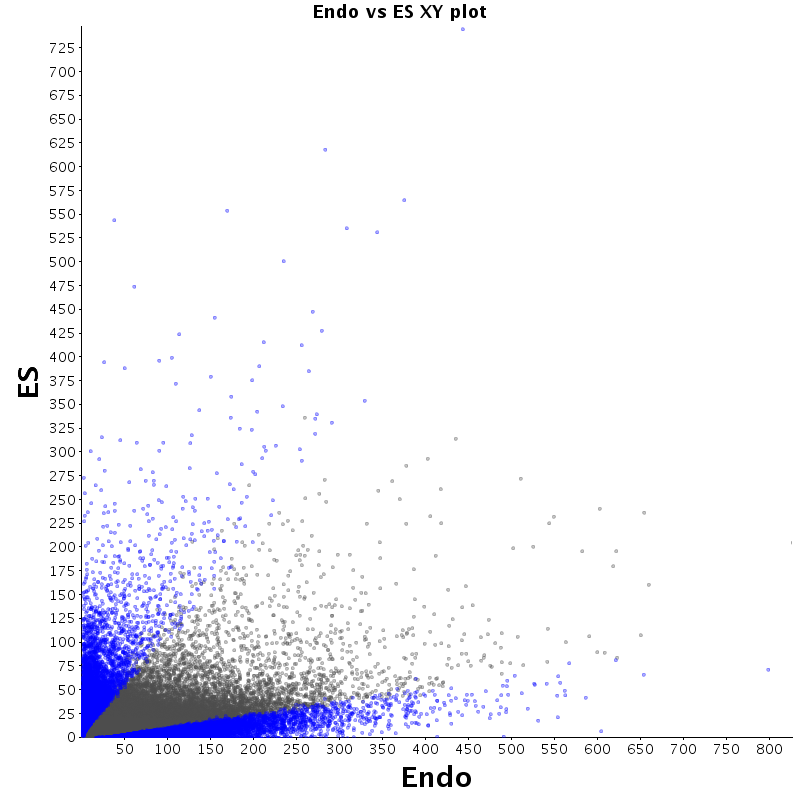 |
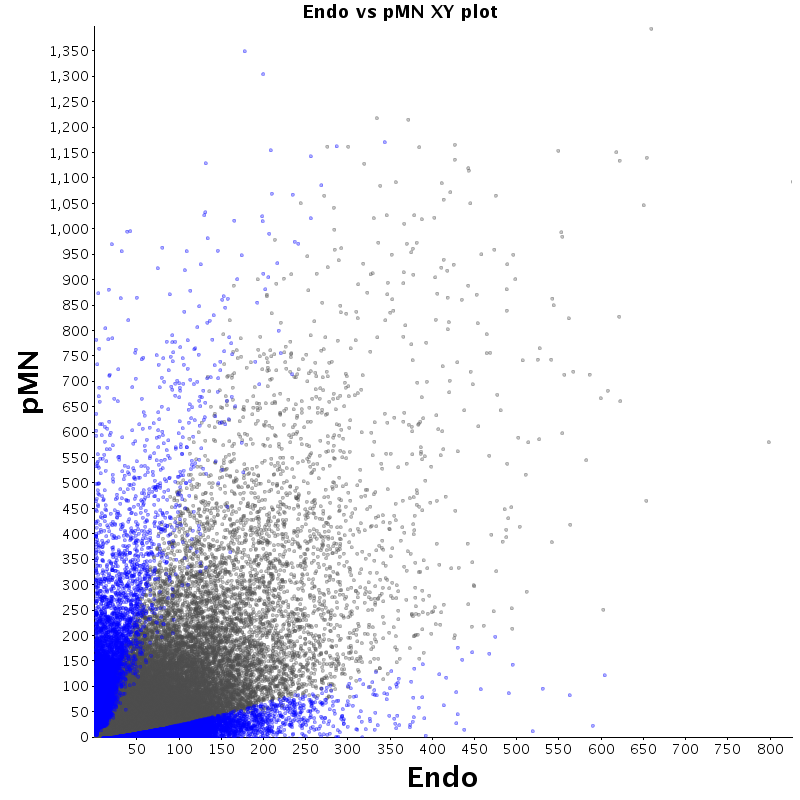 |
| Diff | ES | pMN | Endo |
|---|---|---|---|
| ES | 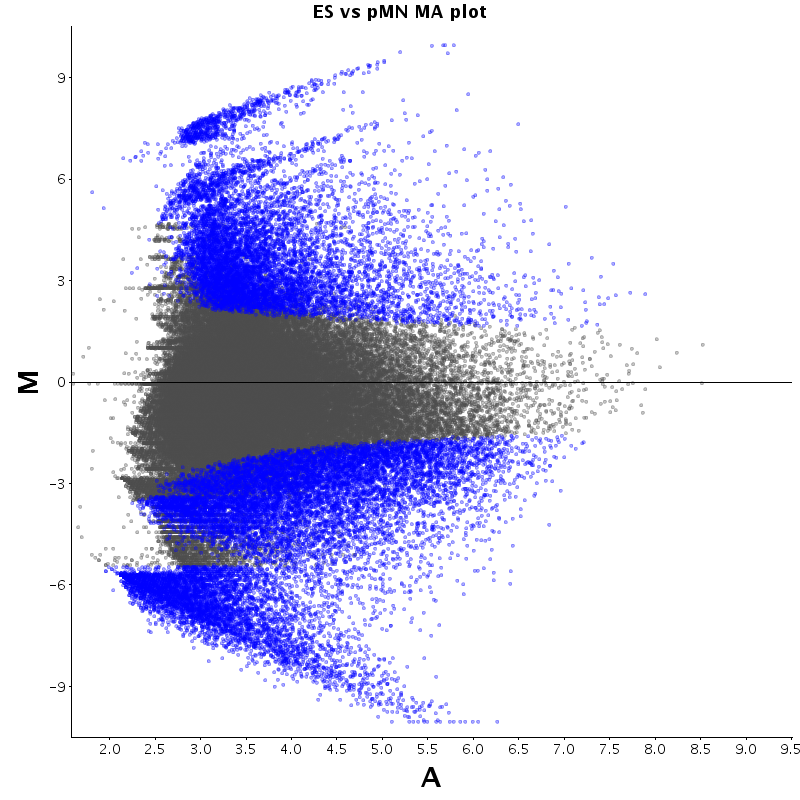 |
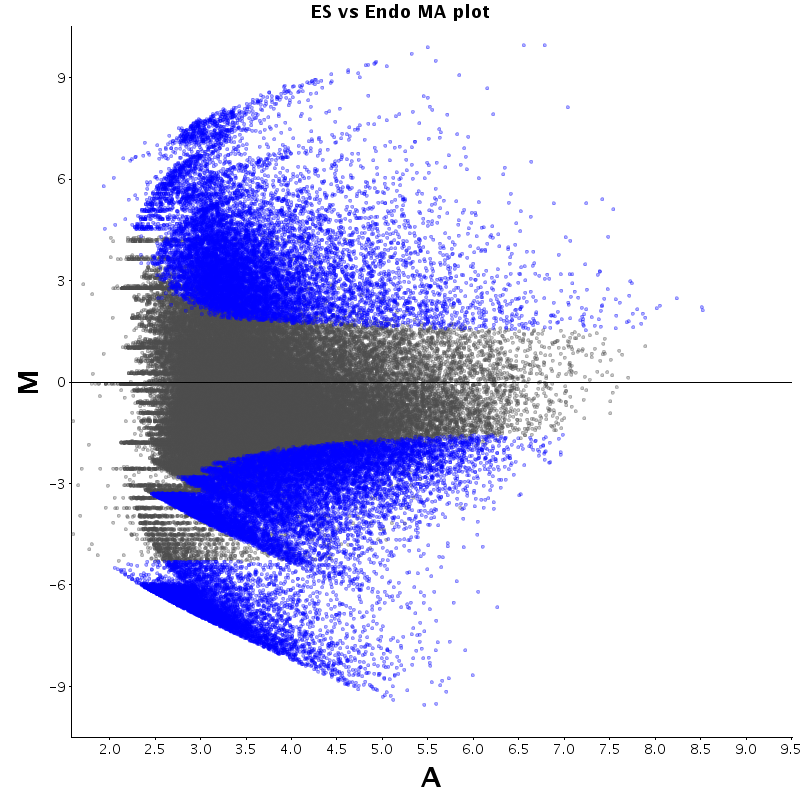 |
|
| pMN | 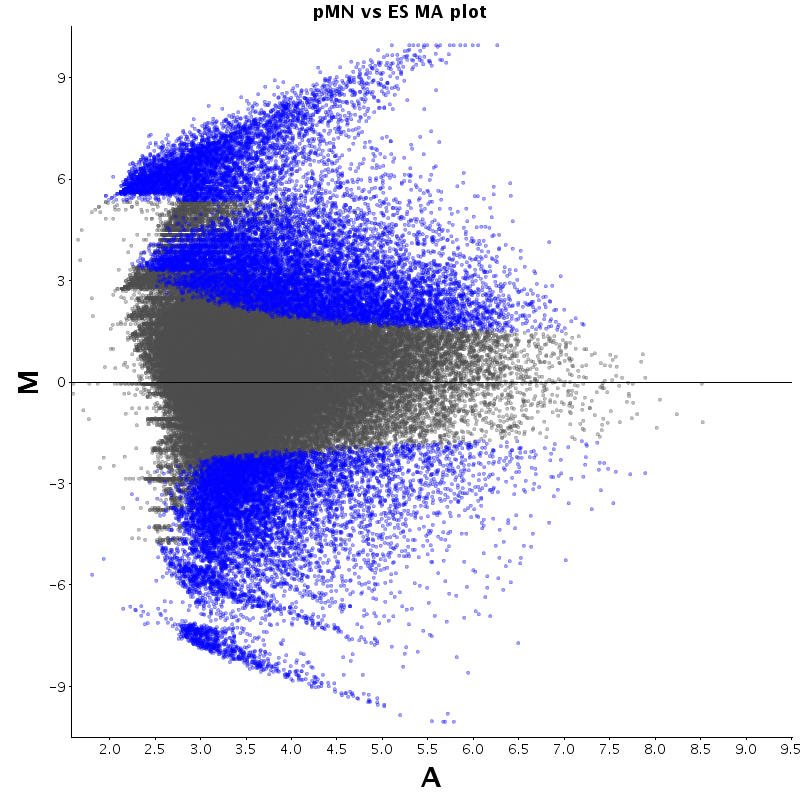 |
 |
|
| Endo | 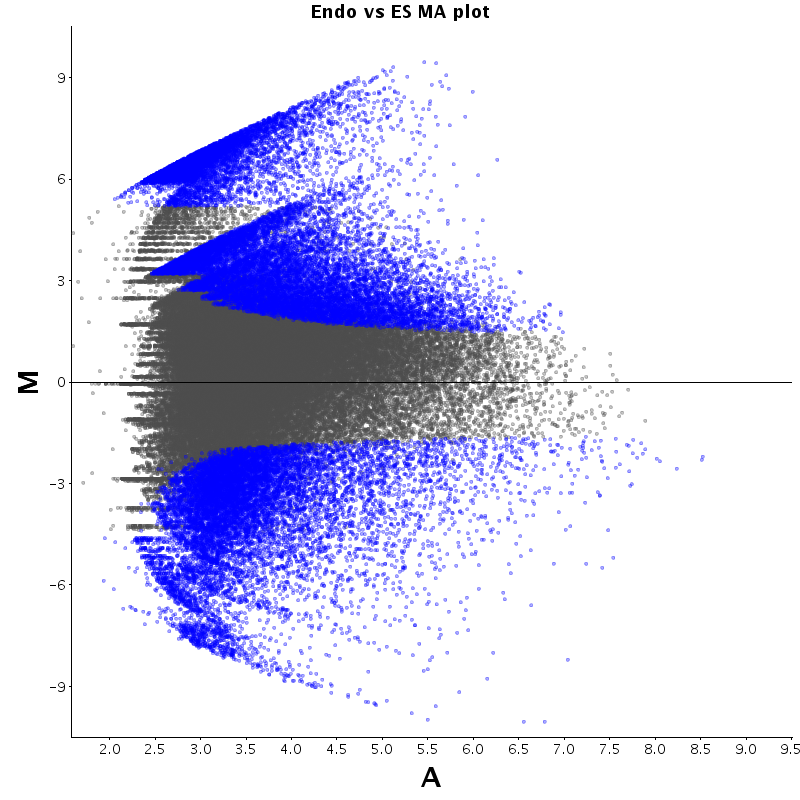 |
 |
Positional prior motifs. Try inputting these motifs into STAMP for validation.
Peak-peak distance histograms (same condition)